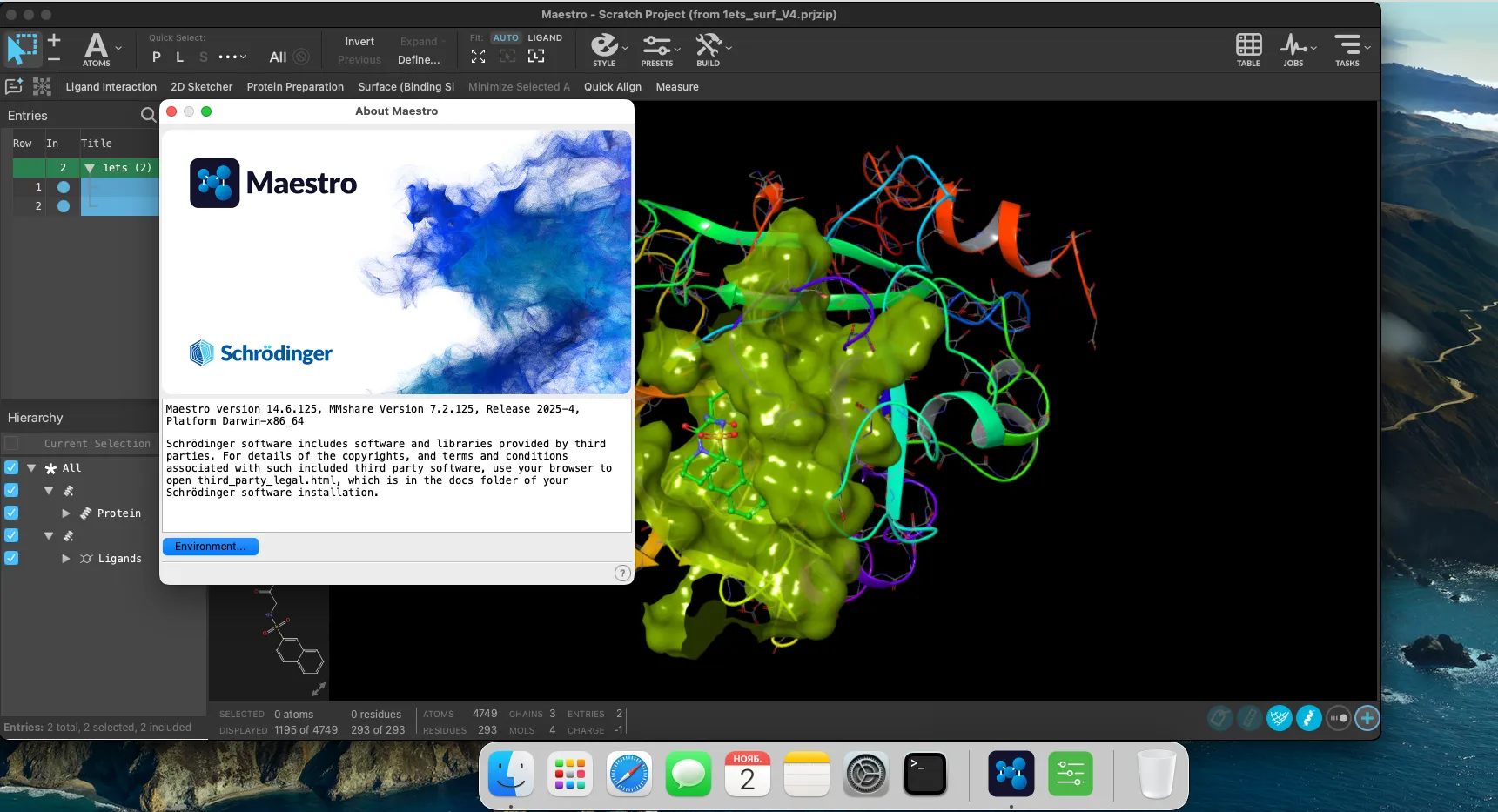
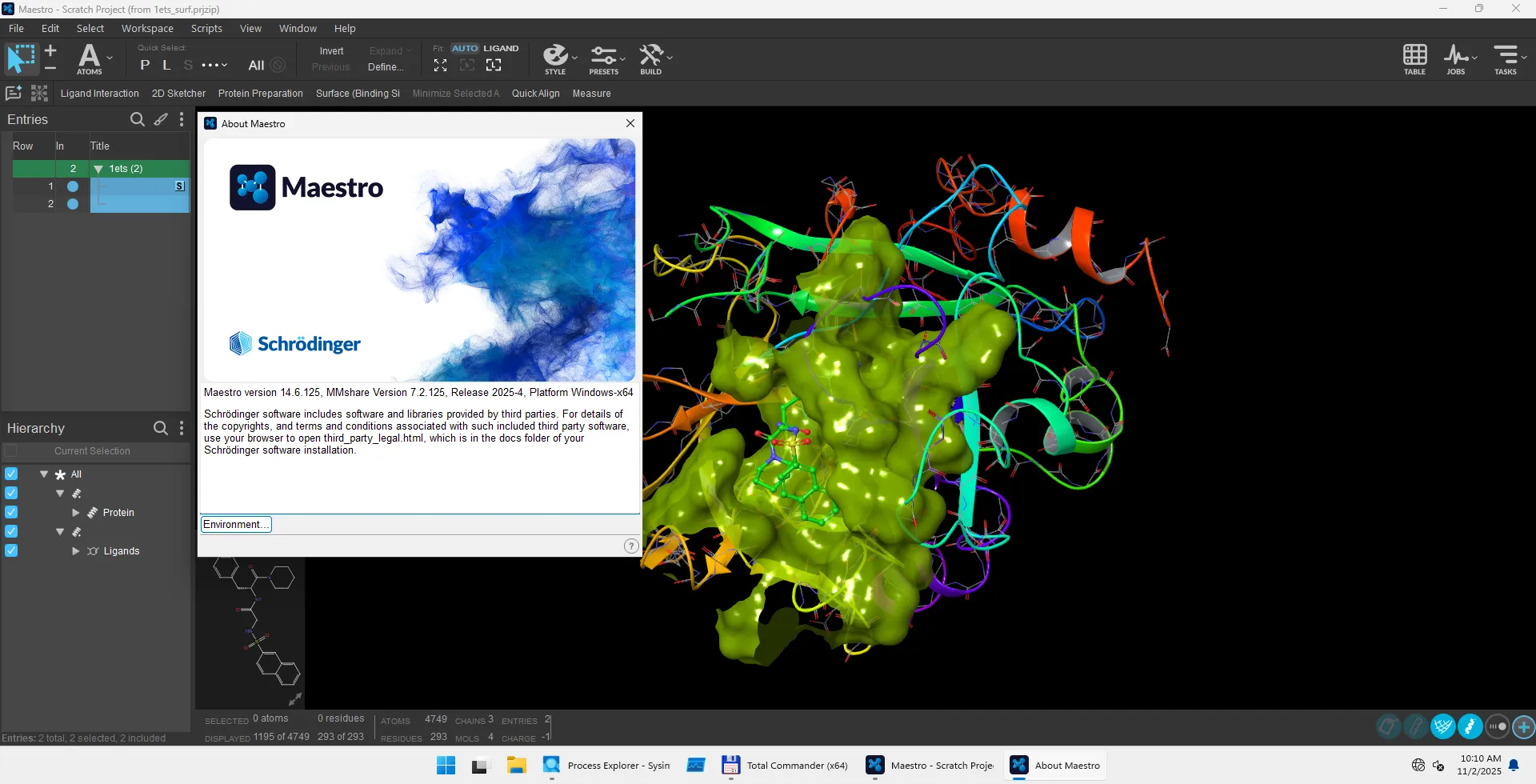
Schrodinger Suites 2025-4 | 30.1 Gb
The software developer Schrödinger continuously strive to develop scientific solutions that push the boundaries of molecular design and are delighted to share enhanced drug discovery and materials workflows in Schrödinger’s 2025-4 software release.
Small Molecule Drug Discovery
Platform Environment
Maestro Graphical Interface
- One-click deletion of empty project table columns: A single menu action instantly removes all empty property columns from the entire Project Table, decluttering the workspace to allow a clearer focus on relevant data.
- “Select Entries in Table” from workspace right-click menu: Right-clicking any atom in the Workspace instantly locates and selects its corresponding entry in the Project Table, eliminating the need for manual searching in large projects.
- Maestro Assistant (Beta) enhancements: Maestro Assistant is now more interactive with the addition of a command history, copy-to-clipboard functionality, clearer mode notifications, and a streamlined feedback system.
Binding Site & Structure Analysis
SiteMap
- SiteMap now evaluates more sites than requested, returning the requested number of top-scoring sites by SiteScore
Desmond Molecular Dynamics
- Energy Decomposition Analysis panel now supports trajectories generated using custom force field parameters
Mixed Solvent MD (MxMD)
- Addition of immiscible probes in Mixed Solvent MD to better identify cryptic pockets: Use hydrophobic probes to stabilize transiently exposed pockets in an open conformation to increase their residence time and facilitate cryptic binding site identification
Ligand Preparation
Pharmacophore Modeling
- Use 1D similarities to screen against massive virtual spaces defined combinatorically with the combinatorial_explorer workflow (command line only)
Lead Optimization
Ligand alignment
- Non macrocycle alignment jobs can run as interactive tasks without submitting a job
Enumeration
- Return only Pathfinder retrosynthetic routes that include purchasable starting materials
FEP+
- Added ligand atomic RMSF analysis for RE-FEP calculations
FEP Protocol Builder
- Enable separate solvent and complex hot atom rules including None
- Enable sampling between OPLS4 and OPLS5 with the new Force Field version parameter
Quantum Mechanics
- Predict optical rotation as a function of wavelength
- Added support for 18 new double hybrid functionals
- Faster simulations with optimized CPU assignments for unbalanced batch jobs and workflows
- Employ QRNN, MPNICE and UMA MLFFs from Jaguar interfaces
Medical Chemistry Design
Ligand Designer
- Interactive Core Hopping: A new Core Swap workflow supports non-ring cores and enables true core hopping transformations, allowing for the exploration of diverse chemical space directly within an interactive design environment.
- Manual attachment point selection for macrocyclization: The newly renamed Macrocyclization workflow allows for the interactive selection of attachment points, providing precise chemical control that focuses linker enumeration on synthetically relevant designs.
De Novo Design
- New PDF report for AutoDesigner workflows to record input settings and summarize enumeration and filtering results
Drug Formulations
Crystal Structure Prediction
- Expansion of drug formulations applicability with highly accurate Z’=2 polymorph predictions (Beta): Identify stable crystal polymorphs with updated crystal structure prediction capabilities to sample, search and rank confidently
Alternative Modalities
Bifunctional Degraders
- Optionally build linkers between the two warheads in both directions when using the “Linkers only” option in the Generate Degrader Ternary Complexes interface
- Prioritize ternary complexes for linker design with a new interface to score degrader ternary complexes powered by a validated Metadynamics-based workflow (beta)
Education Content
- New Learning Path: T Cell Receptor Engineering
- New Tutorial: Enzyme Engineering with BioLuminate
- Updated Tutorial: Validating a Protein Free Energy Perturbation Model for Thermostability Predictions for Single Point Mutations (previously “Obtaining Protein Free Energy Perturbation Thermostability Predictions for Single Point Mutations”)
- Updated Tutorial: Improving the Thermostability of T4 Lysozyme Using Protein FEP+ Guided Design (previously “Introduction to Protein Thermostability Prediction using Protein FEP+”)
- Updated Tutorial: Ligand Binding Pose Generation for FEP+ (previously Ligand Binding Pose Prediction for FEP+ using Core-Constrained Docking)
- Updated Tutorial: Structure-Based Virtual Screening using Glide
Biologics Drug Discovery
- Predict T-Cell Receptor structures using TCRBuilder2 through Maestro (full release)
- Improved ability to visualize sequences before and after CDR grafting in Antibody Humanization Panel by replacing the pop-up sequence viewer with the MSV
- Streamlined visualization and plotting of mutation results by updating the mutations chart in the residue scanning results viewer interface
- Simpler analysis in the Residue Scanning Viewer by synchronization between chart and table
- In MM-GBSA Residue Scanning Viewer interface choose properties to plot on both axis, eg. plot affinity on x and stability on y-axis
- Easier setup of MM-GBSA Residue Scanning calculations through synchronization of selection between workspace and table, automatic fit-on selection, and ability to toggle visibility of select columns in property table
- New ASPmax protein descriptor capturing the maximum average surface property for aggregation propensity prediction (command line)
GUI for Quantum ESPRESSO
Product: Quantum ESPRESSO (QE) Interface
- (+DEFECT_FORMATION_ENERGY) Defect Correction: Workflow solution to compute the defect formation energy
- Upgrade to Quantum ESPRESSO 7.5
- Support for 3-body dispersion correction
- Visualization of d-band center
- Support for finite displacement phonon calculations
- Reduced disk space usage for phonon calculations
- Option to set self-consistency threshold for phonons
MS Surface
Product: MS SurfChem
- Desorption Enumeration: Active entry shown in the workspace
- Desorption Enumeration: Support for enumerating associate desorption products
Microkinetics
Product: MS Microkinetics
- Option to calculate the degree of selectivity control
- Option to plot degree of rate control per species
- Option to set pressure schedule per species
- Setup for transition state lateness parameter to define lateral interactions
- Setup for adsorbate-adsorbate interactions via lateral scaling parameter
- Support for saving and loading reaction network files
- Support for simplified reaction entry using plain text
- Support for loading a microkinetic model from a workspace entry
- Support for plotting x-axis in log scale from the viewer panel
- User control over absolute and relative error tolerances
- Number of unphysical MKM steps displayed in viewer panel
Reactivity
Product: MS Reactivity
- Nanoreactor: Option to set spin for the final state of elementary reactions
- Nanoreactor: Refined setup for metadynamics cavity radius scaled from 0.1 to 1.0
- Nanoreactor: Product chemistry filtered by refined DFT or MLFF
- Nanoreactor: Reactant energy marked in the elementary reaction network mode
- Nanoreactor: Support for MLFF (UMA) energy refinement for open shell systems
- Reaction Network Profiler: Option to sort conformers from refined energy
Advanced Force Field Applications
Product: MS FF Applications
- Expanded support for MLFF selections in QM- and MD-based workflow solutions
- UMA (developed by Meta Platforms Inc.) added as MLFF option in QM-based workflows
- (+ENABLE_GRPC_MLFF_DESMOND) UMA (developed by Meta Platforms Inc.) added as MLFF option in MD-based workflows
Transport Calculations via MD simulations
Product: MS Transport
- Ionic Conductivity: Workflow solution to predict ionic conductivity in liquid electrolyte systems
- Viscosity: Option to turn off the SHAKE algorithm (command line)
- Viscosity: Support for MLFF
Coarse-Grained (CG) Molecular Dynamics
Product: MS CG
- Coarse-grained Mapping: Consistent, reusable names for sugar particles
- Coarse-grained Mapping: Improved restraint visualization
- Coarse-grained Mapping: MARTINI mapping of proteins
- Coarse-grained Mapping: MARTINI mapping of monosaccharides
- Coarse-grained Mapping: MARTINI mapping of cholesterol
- Coarse-grained Mapping: All-atom CMS created as input for CG FF Builder
- Coarse-Grained Mapping: Enhanced precision for SMARTS patterns to improve mapping and force field parameter reusability
- Coarse-Grained Mapping: Improved UX for the reuse of existing particle types
- Coarse-Grained Mapping: Visualization of the mapped system in the workspace
- Coarse-Grained Mapping: Use of antifreeze water molecules set by default
- CG FF Builder: NpT set as the default ensemble for MARTINI simulations
- CG FF Builder: Option to keep proteins rigid during the model building stage
- CG FF Builder: Option to set the masses to standard MARTINI particle types
- CG FF Builder: Mapping of a small ion-water cluster to a single MARTINI particle
- CG FF Builder: Support for the reuse of CG particle types with identical names
- CG FF Builder: Support for loading CG mapping output as input
Complex Bilayer Builder
Product: MS Complex Bilayer Builder
- Complex Bilayer: Model building solution for complex / multi-component bilayers of molecular materials including protein-based membranes
- Membrane Analysis: Workflow solution to analyze membrane structural features
Formulation ML
Product: MS Formulation ML
- Formulation ML: Support for parallel training and predictions of multiple models
- Formulation ML Optimization: Improved UX for loading models
- Formulation ML Optimization: Option to select random optimization for models
- Formulation ML Optimization: Option to stop optimization prior to convergence
- Formulation ML Optimization: Option for cost optimization
- Formulation ML Optimization: Support for composition constraints with Bayesian optimization
- ML Model Manager: Option to export and update descriptors for ingredients
- ML Model Manager: Automatic selection for the newly loaded model
- ML Model Manager: Access for model names to be edited by user
- ML Model Manager: Option to estimate MPO scores on model predictions
Layered Device ML
Product: MS Layered Device ML
- OLED Device ML: Advanced options for model training
- OLED Device ML: Option to export the training set data
- OLED Device ML: Support for parallel training and predictions of multiple models
- OLED Device ML: Option to use molecular model predictions as descriptors
MS Maestro User Interface
- Maestro: Job Monitor to display the cause of failures for failed jobs
- Maestro: Ribbon style enabled in the workspace for protein representations
MS Maestro Builders and Tools
- Complex Builder: Option to turn on/off IUPAC name assignment for ligands
- Complex Builder: Support for building dimers with an atom bridging two metals
- Disordered System: Preservation of protein residue information by default
- Disordered System: Option to set the system size by the total number of atoms
- Materials Science Panel Explorer: GUI to search and list panels by the application, method, chemistry, and product of interest
- Meta Workflows: Matched settings for the Brownie stage with the MD Multistage
- Optoelectronic Device Designer: Option to import materials data from a file
- Optoelectronic Device Designer: Option to export materials and device data
- Optoelectronic Device Designer: Option to remove materials from the database
- Optoelectronic Device Designer: Plot for numerical energy levels
- Optoelectronic Device Designer: Plot for numerical layer thicknesses
- Solvate System: Option to specify particle radii for coarse-grained models
- Solvate System, Structured Liquid GUI: (+MATSCI_PACKMOL_PBC) Periodic boundary conditions retained when building structures
Classical Mechanics
- Cluster Analysis: (+CLUSTER_DENSITY_PROFILE) Radial density profile for clusters
- Evaporation: Support for MLFF
- Evaporation: Setup for evaporation zone in radial distance from center of mass
- Polymer Crosslink: (+POLYMER_CROSSLINK_MODES) Option to select fast crosslinking mode
- Thin Plane Shear: Support for MLFF
- Thin Plane Shear: Option to use custom MLFF
- Umbrella Sampling: User control over potential of mean force (PMF) calculations
- Umbrella Sampling: Visualization of probability distribution overlap matrix
- Visualize Restraints: Visualization of multiple restraints
Quantum Mechanics
- Adsorption Site Finder: Support for MLFF
- Bond and Ligand Dissociation: Support for MLFF
- QM Multistage: Support for MLFF selection on the Theory tab
- Optoelectronic Film Properties: Advanced transition dipole moment analysis with distributions over angle, distance, and depth
- Probe Grid Scan: VdW radius used as atomic radius for metal atoms
- Probe Grid Scan: Support for scanning open shell systems using MLFF (UMA)
Education Content
- New Tutorial: Simulating Complex Protein Solutions
- New Tutorial: Creating a Coarse-Grained Model for Protein Formulations
- New Tutorial: Building and Analyzing a Complex Lipid Bilayer and Embedding a Membrane Protein
- New Tutorial: Ionic Conductivity
- New Tutorial: Optimizing Viscosity and Cost in Formulations with Missing Structural Data
- New Tutorial: Locating Adsorption Sites on Surfaces
- Updated Tutorial: Atomic Layer Deposition
- Updated Tutorial: Microkinetic Modeling
- Quick Reference Sheet: Materials Science Panel Explorer
Education Content
Life Science
- New Learning Path: T Cell Receptor Engineering
- New Tutorial: Enzyme Engineering with BioLuminate
- Updated Tutorial: Validating a Protein Free Energy Perturbation Model for Thermostability Predictions for Single Point Mutations (previously “Obtaining Protein Free Energy Perturbation Thermostability Predictions for Single Point Mutations”)
- Updated Tutorial: Improving the Thermostability of T4 Lysozyme Using Protein FEP+ Guided Design (previously “Introduction to Protein Thermostability Prediction using Protein FEP+”)
- Updated Tutorial: Ligand Binding Pose Generation for FEP+ (previously Ligand Binding Pose Prediction for FEP+ using Core-Constrained Docking)
- Updated Tutorial: Structure-Based Virtual Screening using Glide
Materials Science
- New Tutorial: Simulating Complex Protein Solutions
- New Tutorial: Creating a Coarse-Grained Model for Protein Formulations
- New Tutorial: Building and Analyzing a Complex Lipid Bilayer and Embedding a Membrane Protein
- New Tutorial: Ionic Conductivity
- New Tutorial: Optimizing Viscosity and Cost in Formulations with Missing Structural Data
- New Tutorial: Locating Adsorption Sites on Surfaces
- Updated Tutorial: Atomic Layer Deposition
- Updated Tutorial: Microkinetic Modeling
- Quick Reference Sheet: Materials Science Panel Explorer
Schrödinger Software provide accurate, reliable, and high performance computational technology to solve real-world problems in life science research. It can be used to build, edit, run and analyse molecules. The Schrödinger-Suite of applications have a graphical user interface called Maestro. Using the Maestro you can prepare your structure for refinement. The following products are available: CombiClide, ConfGen, Core Hopping, Desmond, Epik, Glide, Glide, Impact, Jaguar ( high-performance ab initio package), Liaison, LigPrep, MacroModel, MCPRO+, Phase, Prime, PrimeX, QikProp, QSite, Semi-Empirical, SiteMap, and Strike.
Schrödinger Release 2025-4 | Life Science - New Features
Schrödinger Release 2025-4 | Materials Science - New Features
Schrödinger, LLC provides scientific software solutions and services for life sciences and materials research, as well as academic, government, and non-profit institutions around the world. It offers small-molecule drug discovery, biologics, materials science, and discovery informatics solutions; and PyMOL, a 3D molecular visualization solution. The company was founded in 1990 and is based in Portland, Oregon with operations in the United States, Europe, Japan, and India.
Owner: Schrödinger, LLC
Product Name: Schrödinger Suites
Version: 2025-4 Commercial Version *
Supported Architectures: x64
Website Home Page : www.schrodinger.com
Languages Supported: english
System Requirements: Windows, Linux, macOs **
Size: 30.1 Gb
* Notes: All workflows that rely on Desmond are not supported on Windows or Mac platforms, they can only be run on Linux. This includes Molecular Dynamics, IFD-MD, FEP+, WaterMap, and a number of Materials Science workflows.
GPU machine learning applications such as Active Learning Glide and DeepAutoQSAR on GPU can only be run on Linux.
Please visit my blog
Added by 3% of the overall size of the archive of information for the restoration
No mirrors please
Added by 3% of the overall size of the archive of information for the restoration
No mirrors please